Graph classes#
|
The basic graph class, which inherits from a library class such as |
|
The detailed class that inherits from |
|
The detailed class that inherits from |
|
Class that inherits from |
Details#
- class nngt.Graph(*args, **kwargs)[source]#
The basic graph class, which inherits from a library class such as
graph_tool.Graph,networkx.DiGraph, origraph.Graph.The objects provides several functions to easily access some basic properties.
Initialize Graph instance
Changed in version 2.0: Renamed from_graph to copy_graph.
Changed in version 2.2: Added structure argument.
- Parameters
nodes (int, optional (default: 0)) – Number of nodes in the graph.
name (string, optional (default: “Graph”)) – The name of this
Graphinstance.weighted (bool, optional (default: True)) – Whether the graph edges have weight properties.
directed (bool, optional (default: True)) – Whether the graph is directed or undirected.
copy_graph (
Graph, optional) – An optionalGraphthat will be copied.structure (
Structure, optional (default: None)) – A structure dividing the graph into specific groups, which can be used to generate specific connectivities and visualise the connections in a more coarse-grained manner.kwargs (optional keywords arguments) – Optional arguments that can be passed to the graph, e.g. a dict containing information on the synaptic weights (
weights={"distribution": "constant", "value": 2.3}which is equivalent toweights=2.3), the synaptic delays, or atypeinformation.
Note
When using copy_graph, only the topological properties are copied (nodes, edges, and attributes), spatial and biological properties are ignored. To copy a graph exactly, use
copy().- Returns
self (
Graph)
- adjacency_matrix(types=False, weights=False, mformat='csr')[source]#
Return the graph adjacency matrix.
Note
Source nodes are represented by the rows, targets by the corresponding columns.
- Parameters
types (bool, optional (default: False)) – Wether the edge types should be taken into account (negative values for inhibitory connections).
weights (bool or string, optional (default: False)) – Whether the adjacecy matrix should be weighted. If True, all connections are multiply bythe associated synaptic strength; if weight is a string, the connections are scaled bythe corresponding edge attribute.
mformat (str, optional (default: “csr”)) – Type of
scipy.sparsematrix that will be returned, by defaultscipy.sparse.csr_matrix.
- Returns
mat (
scipy.sparsematrix) – The adjacency matrix of the graph.
- clear_all_edges()#
Remove all edges from the graph
- delete_edges(edges)#
Remove a list of edges
- delete_nodes(nodes)#
Remove nodes (and associated edges) from the graph.
- property edge_attributes#
Access edge attributes.
- edge_id(edge)#
Return the ID a given edge or a list of edges in the graph. Raises an error if the edge is not in the graph or if one of the vertices in the edge is nonexistent.
- Parameters
edge (2-tuple or array of edges) – Edge descriptor (source, target).
- Returns
index (int or array of ints) – Index of the given edge.
- edge_nb()#
Number of edges in the graph
- property edges_array#
Edges of the graph, sorted by order of creation, as an array of 2-tuple.
- static from_file(filename, fmt='auto', separator=' ', secondary=';', attributes=None, attributes_types=None, notifier='@', ignore='#', from_string=False, name=None, directed=True, cleanup=False)[source]#
Import a saved graph from a file.
Changed in version 2.0: Added optional attributes_types and cleanup arguments.
- Parameters
filename (str) – The path to the file.
fmt (str, optional (default: deduced from filename)) – The format used to save the graph. Supported formats are: “neighbour” (neighbour list), “ssp” (scipy.sparse), “edge_list” (list of all the edges in the graph, one edge per line, represented by a
source target-pair), “gml” (gml format, default if filename ends with ‘.gml’), “graphml” (graphml format, default if filename ends with ‘.graphml’ or ‘.xml’), “dot” (dot format, default if filename ends with ‘.dot’), “gt” (only when using graph_tool as library, detected if filename ends with ‘.gt’).separator (str, optional (default ” “)) – separator used to separate inputs in the case of custom formats (namely “neighbour” and “edge_list”)
secondary (str, optional (default: “;”)) – Secondary separator used to separate attributes in the case of custom formats.
attributes (list, optional (default: [])) – List of names for the attributes present in the file. If a notifier is present in the file, names will be deduced from it; otherwise the attributes will be numbered. For “edge_list”, attributes may also be present as additional columns after the source and the target.
attributes_types (dict, optional (default: str)) – Backup information if the type of the attributes is not specified in the file. Values must be callables (types or functions) that will take the argument value as a string input and convert it to the proper type.
notifier (str, optional (default: “@”)) – Symbol specifying the following as meaningfull information. Relevant information are formatted
@info_name=info_value, whereinfo_nameis in (“attributes”, “directed”, “name”, “size”) and associatedinfo_valueare of type (list,bool,str,int). Additional notifiers are@type=SpatialGraph/Network/SpatialNetwork, which must be followed by the relevant notifiers among@shape,@population, and@graph.from_string (bool, optional (default: False)) – Load from a string instead of a file.
ignore (str, optional (default: “#”)) – Ignore lines starting with the ignore string.
name (str, optional (default: from file information or ‘LoadedGraph’)) – The name of the graph.
directed (bool, optional (default: from file information or True)) – Whether the graph is directed or not.
cleanup (bool, optional (default: False)) – If true, removes nodes before the first one that appears in the edges and after the last one and renumber the nodes from 0.
- Returns
graph (
Graphor subclass) – Loaded graph.
- classmethod from_library(library_graph, name='ImportedGraph', weighted=True, directed=True, **kwargs)[source]#
Create a
Graphby wrapping a graph object from one of the supported libraries.- Parameters
library_graph (object) – Graph object from one of the supported libraries (graph-tool, igraph, networkx).
name (str, optional (default: “ImportedGraph”))
**kwargs – Other standard arguments (see
__init__())
- classmethod from_matrix(matrix, weighted=True, directed=True, population=None, shape=None, positions=None, name=None, **kwargs)[source]#
Creates a
Graphfrom ascipy.sparsematrix or a dense matrix.- Parameters
matrix (
scipy.sparsematrix ornumpy.ndarray) – Adjacency matrix.weighted (bool, optional (default: True)) – Whether the graph edges have weight properties.
directed (bool, optional (default: True)) – Whether the graph is directed or undirected.
population (
NeuralPop) – Population to associate to the newNetwork.shape (
Shape, optional (default: None)) – Shape to associate to the newSpatialGraph.positions ((N, 2) array) – Positions, in a 2D space, of the N neurons.
name (str, optional) – Graph name.
- Returns
- get_attribute_type(attribute_name, attribute_class=None)[source]#
Return the type of an attribute (e.g. string, double, int).
- Parameters
attribute_name (str) – Name of the attribute.
attribute_class (str, optional (default: both)) – Whether attribute_name is a “node” or an “edge” attribute.
- Returns
type (str) – Type of the attribute.
- get_betweenness(btype='both', weights=None)[source]#
Returns the normalized betweenness centrality of the nodes and edges.
- Parameters
g (
Graph) – Graph to analyze.btype (str, optional (default ‘both’)) – The centrality that should be returned (either ‘node’, ‘edge’, or ‘both’). By default, both betweenness centralities are computed.
weights (bool or str, optional (default: binary edges)) – Whether edge weights should be considered; if
NoneorFalsethen use binary edges; ifTrue, uses the ‘weight’ edge attribute, otherwise uses any valid edge attribute required.
- Returns
nb (
numpy.ndarray) – The nodes’ betweenness if btype is ‘node’ or ‘both’eb (
numpy.ndarray) – The edges’ betweenness if btype is ‘edge’ or ‘both’
See also
- get_degrees(mode='total', nodes=None, weights=None, edge_type='all')[source]#
Degree sequence of all the nodes.
Changed in version 2.0: Changed deg_type to mode, node_list to nodes, use_weights to weights, and edge_type to edge_type.
- Parameters
mode (string, optional (default: “total”)) – Degree type (among ‘in’, ‘out’ or ‘total’).
nodes (list, optional (default: None)) – List of the nodes which degree should be returned
weights (bool or str, optional (default: binary edges)) – Whether edge weights should be considered; if
NoneorFalsethen use binary edges; ifTrue, uses the ‘weight’ edge attribute, otherwise uses any valid edge attribute required.edge_type (int or str, optional (default: all)) – Restrict to a given synaptic type (“excitatory”, 1, or “inhibitory”, -1), using either the “type” edge attribute for non-
Networkor theinhibitorynodes.
- Returns
degrees (
numpy.array).. warning :: – When using MPI with “nngt” (distributed) backend, returns only the degrees associated to local edges. “Complete” degrees are obtained by taking the sum of the results on all MPI processes.
- get_delays(edges=None)[source]#
Returns the delays of all or a subset of the edges.
Changed in version 1.0.1: Added the possibility to ask for a subset of edges.
- Parameters
edges ((E, 2) array, optional (default: all edges)) – Edges for which the type should be returned.
- Returns
the list of delays
- get_density()[source]#
Density of the graph:
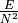 , where E is the number
of edges and N the number of nodes.
, where E is the number
of edges and N the number of nodes.
- get_edge_attributes(edges=None, name=None)[source]#
Attributes of the graph’s edges.
Changed in version 1.0: Returns the full dict of edges attributes if called without arguments.
New in version 0.8.
- Parameters
edge (tuple or list of tuples, optional (default:
None)) – Edge whose attribute should be displayed.name (str, optional (default:
None)) – Name of the desired attribute.
- Returns
Dict containing all graph’s attributes (synaptic weights, delays…)
by default. If edge is specified, returns only the values for these
edges. If name is specified, returns value of the attribute for each
edge.
Note
The attributes values are ordered as the edges in
edges_array()if edges is None.
- get_edge_types(edges=None)[source]#
Return the type of all or a subset of the edges.
- Parameters
edges ((E, 2) array, optional (default: all edges)) – Edges for which the type should be returned.
- Returns
the list of types (1 for excitatory, -1 for inhibitory)
- get_edges(attribute=None, value=None, source_node=None, target_node=None)[source]#
Return the edges in the network fulfilling a given condition.
For undirected graphs, edges are always returned in the order
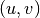 where
where 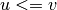 .
.Warning
Contrary to
edges_array()that returns edges ordered by creation time (i.e. corresponding to the order of the edge attribute array), this function does not enforce any specific edge order. This also means that, if order does not matter, it may be faster to callget_edgesthat to calledges_array.- Parameters
attribute (str, optional (default: all nodes)) – Whether the attribute of the returned edges should have a specific value.
value (object, optional (default : None)) – If an attribute name is passed, then only edges with attribute being equal to value will be returned.
source_node (int or list of ints, optional (default: all nodes)) – Retrict the edges to those stemming from source_node.
target_node (int or list of ints, optional (default: all nodes)) – Retrict the edges to those arriving at target_node.
- Returns
A list of edges (2-tuples).
See also
- get_node_attributes(nodes=None, name=None)[source]#
Attributes of the graph’s edges.
Changed in version 1.0.1: Corrected default behavior and made it the same as
get_edge_attributes().New in version 0.9.
- Parameters
nodes (list of ints, optional (default:
None)) – Nodes whose attribute should be displayed.name (str, optional (default:
None)) – Name of the desired attribute.
- Returns
Dict containing all nodes attributes by default. If nodes is
specified, returns a
dictcontaining only the attributes of thesenodes. If name is specified, returns a list containing the values of
the specific attribute for the required nodes (or all nodes if
unspecified).
See also
get_edge_attributes(),new_node_attribute(),set_node_attribute(),new_edge_attributes(),set_edge_attribute()
- get_nodes(attribute=None, value=None)[source]#
Return the nodes in the network fulfilling a given condition.
- Parameters
attribute (str, optional (default: all nodes)) – Whether the attribute of the returned nodes should have a specific value.
value (object, optional (default : None)) – If an attribute name is passed, then only nodes with attribute being equal to value will be returned.
See also
- get_structure_graph()[source]#
Return a coarse-grained version of the graph containing one node per
nngt.Group. Connections between groups are associated to the sum of all connection weights. If no structure is present, returns an empty Graph.
- get_weights(edges=None)[source]#
Returns the weights of all or a subset of the edges.
Changed in version 1.0.1: Added the possibility to ask for a subset of edges.
- Parameters
edges ((E, 2) array, optional (default: all edges)) – Edges for which the type should be returned.
- Returns
the list of weights
- property graph#
Returns the underlying library object.
Warning
Do not add or remove edges directly through this object.
- is_connected(mode='strong')[source]#
Return whether the graph is connected.
- Parameters
mode (str, optional (default: “strong”)) – Whether to test connectedness with directed (“strong”) or undirected (“weak”) connections.
References
- ig-connected
- is_network()[source]#
Whether the graph is a subclass of
Network(i.e. if it has aNeuralPopattribute).
- is_spatial()[source]#
Whether the graph is embedded in space (i.e. is a subclass of
SpatialGraph).
- static make_network(graph, neural_pop, copy=False, **kwargs)[source]#
Turn a
Graphobject into aNetwork, or aSpatialGraphinto aSpatialNetwork.- Parameters
graph (
GraphorSpatialGraph) – Graph to convertneural_pop (
NeuralPop) – Population to associate to the newNetworkcopy (bool, optional (default:
False)) – Whether the operation should be made in-place on the object or if a new object should be returned.
Notes
In-place operation that directly converts the original graph if copy is
False, else returns the copiedGraphturned into aNetwork.
- static make_spatial(graph, shape=None, positions=None, copy=False)[source]#
Turn a
Graphobject into aSpatialGraph, or aNetworkinto aSpatialNetwork.- Parameters
graph (
GraphorSpatialGraph) – Graph to convert.shape (
Shape, optional (default: None)) – Shape to associate to the newSpatialGraph.positions ((N, 2) array) – Positions, in a 2D space, of the N neurons.
copy (bool, optional (default:
False)) – Whether the operation should be made in-place on the object or if a new object should be returned.
Notes
In-place operation that directly converts the original graph if copy is
False, else returns the copiedGraphturned into aSpatialGraph. The shape argument can be skipped if positions are given; in that case, the neurons will be embedded in a rectangle that contains them all.
- property name#
Name of the graph.
- neighbours(node, mode='all')[source]#
Return the neighbours of node.
- Parameters
node (int) – Index of the node of interest.
mode (string, optional (default: “all”)) – Type of neighbours that will be returned: “all” returns all the neighbours regardless of directionality, “in” returns the in-neighbours (also called predecessors) and “out” retruns the out-neighbours (or successors).
- Returns
neighbours (set) – The neighbours of node.
- new_edge(source, target, attributes=None, ignore=False, self_loop=False)#
Adding a connection to the graph, with optional properties.
Changed in version 2.0: Added self_loop argument to enable adding self-loops.
- Parameters
source (
int/node) – Source node.target (
int/node) – Target node.attributes (
dict, optional (default:{})) – Dictionary containing optional edge properties. If the graph is weighted, defaults to{"weight": 1.}, the unit weight for the connection (synaptic strength in NEST).ignore (bool, optional (default: False)) – If set to True, ignore attempts to add an existing edge and accept self-loops; otherwise an error is raised.
self_loop (bool, optional (default: False)) – Whether to allow self-loops or not.
- Returns
The new connection or None if nothing was added.
- new_edge_attribute(name, value_type, values=None, val=None)[source]#
Create a new attribute for the edges.
- Parameters
name (str) – The name of the new attribute.
value_type (str) – Type of the attribute, among ‘int’, ‘double’, ‘string’, or ‘object’
values (array, optional (default: None)) – Values with which the edge attribute should be initialized. (must have one entry per node in the graph)
val (int, float or str , optional (default: None)) – Identical value for all edges.
- new_edges(edge_list, attributes=None, check_duplicates=False, check_self_loops=True, check_existing=True, ignore_invalid=False)#
Add a list of edges to the graph.
Changed in version 2.0: Can perform all possible checks before adding new edges via the
check_duplicatescheck_self_loops, andcheck_existingarguments.- Parameters
edge_list (list of 2-tuples or np.array of shape (edge_nb, 2)) – List of the edges that should be added as tuples (source, target)
attributes (
dict, optional (default:{})) – Dictionary containing optional edge properties. If the graph is weighted, defaults to{"weight": ones}, whereonesis an array the same length as the edge_list containing a unit weight for each connection (synaptic strength in NEST).check_duplicates (bool, optional (default: False)) – Check for duplicate edges within edge_list.
check_self_loops (bool, optional (default: True)) – Check for self-loops.
check_existing (bool, optional (default: True)) – Check whether some of the edges in edge_list already exist in the graph or exist multiple times in edge_list (also performs check_duplicates).
ignore_invalid (bool, optional (default: False)) – Ignore invalid edges: they are not added to the graph and are silently dropped. Unless this is set to true, an error is raised whenever one of the three checks fails.
.. warning:: – Setting check_existing to False will lead to undefined behavior if existing edges are provided! Only use it (for speedup) if you are sure that you are indeed only adding new edges.
- Returns
Returns new edges only.
- new_node(n=1, neuron_type=1, attributes=None, value_types=None, positions=None, groups=None)#
Adding a node to the graph, with optional properties.
- Parameters
n (int, optional (default: 1)) – Number of nodes to add.
neuron_type (int, optional (default: 1)) – Type of neuron (1 for excitatory, -1 for inhibitory)
attributes (dict, optional (default: None)) – Dictionary containing the attributes of the nodes.
value_types (dict, optional (default: None)) – Dict of the attributes types, necessary only if the attributes do not exist yet.
positions (array of shape (n, 2), optional (default: None)) – Positions of the neurons. Valid only for
SpatialGraphorSpatialNetwork.groups (str, int, or list, optional (default: None)) –
NeuralGroupto which the neurons belong. Valid only forNetworkorSpatialNetwork.
- Returns
The node or a list of the nodes created.
- new_node_attribute(name, value_type, values=None, val=None)[source]#
Create a new attribute for the nodes.
- Parameters
name (str) – The name of the new attribute.
value_type (str) – Type of the attribute, among ‘int’, ‘double’, ‘string’, or ‘object’
values (array, optional (default: None)) – Values with which the node attribute should be initialized. (must have one entry per node in the graph)
val (int, float or str , optional (default: None)) – Identical value for all nodes.
- property node_attributes#
Access node attributes.
- node_nb()#
Number of nodes in the graph
- set_delays(delay=None, elist=None, distribution=None, parameters=None, noise_scale=None)[source]#
Set the delay for spike propagation between neurons.
- Parameters
delay (float or class:numpy.array, optional (default: None)) – Value or list of delays (for user defined delays).
elist (class:numpy.array, optional (default: None)) – List of the edges (for user defined delays).
distribution (class:string, optional (default: None)) – Type of distribution (choose among “constant”, “uniform”, “gaussian”, “lognormal”, “lin_corr”, “log_corr”).
parameters (dict, optional (default: {})) – Dictionary containing the properties of the delay distribution.
noise_scale (class:int, optional (default: None)) – Scale of the multiplicative Gaussian noise that should be applied on the delays.
- set_edge_attribute(attribute, values=None, val=None, value_type=None, edges=None)[source]#
Set attributes to the connections between neurons.
Warning
The special “type” attribute cannot be modified when using graphs that inherit from the
Networkclass. This is because for biological networks, neurons make only one kind of synapse, which is determined by thenngt.NeuralGroupthey belong to.- Parameters
attribute (str) – The name of the attribute.
value_type (str) – Type of the attribute, among ‘int’, ‘double’, ‘string’
values (array, optional (default: None)) – Values with which the edge attribute should be initialized. (must have one entry per node in the graph)
val (int, float or str , optional (default: None)) – Identical value for all edges.
value_type (str, optional (default: None)) – Type of the attribute, among ‘int’, ‘double’, ‘string’. Only used if the attribute does not exist and must be created.
edges (list of edges or array of shape (E, 2), optional (default: all)) – Edges whose attributes should be set. Others will remain unchanged.
- set_node_attribute(attribute, values=None, val=None, value_type=None, nodes=None)[source]#
Set attributes to the connections between neurons.
- Parameters
attribute (str) – The name of the attribute.
value_type (str) – Type of the attribute, among ‘int’, ‘double’, ‘string’
values (array, optional (default: None)) – Values with which the edge attribute should be initialized. (must have one entry per node in the graph)
val (int, float or str , optional (default: None)) – Identical value for all edges.
value_type (str, optional (default: None)) – Type of the attribute, among ‘int’, ‘double’, ‘string’. Only used if the attribute does not exist and must be created.
nodes (list of nodes, optional (default: all)) – Nodes whose attributes should be set. Others will remain unchanged.
- set_types(edge_type, nodes=None, fraction=None)[source]#
Set the synaptic/connection types.
Changed in version 2.0: Changed syn_type to edge_type.
Warning
The special “type” attribute cannot be modified when using graphs that inherit from the
Networkclass. This is because for biological networks, neurons make only one kind of synapse, which is determined by thenngt.NeuralGroupthey belong to.- Parameters
edge_type (int, string, or array of ints) – Type of the connection among ‘excitatory’ (also 1) or ‘inhibitory’ (also -1).
nodes (int, float or list, optional (default: None)) – If nodes is an int, number of nodes of the required type that will be created in the graph (all connections from inhibitory nodes are inhibitory); if it is a float, ratio of edge_type nodes in the graph; if it is a list, ids of the edge_type nodes.
fraction (float, optional (default: None)) – Fraction of the selected edges that will be set as edge_type (if nodes is not None, it is the fraction of the specified nodes’ edges, otherwise it is the fraction of all edges in the graph).
- Returns
t_list (
numpy.ndarray) – List of the types in an order that matches the edges attribute of the graph.
- set_weights(weight=None, elist=None, distribution=None, parameters=None, noise_scale=None)[source]#
Set the synaptic weights.
- Parameters
weight (float or class:numpy.array, optional (default: None)) – Value or list of the weights (for user defined weights).
elist (class:numpy.array, optional (default: None)) – List of the edges (for user defined weights).
distribution (class:string, optional (default: None)) – Type of distribution (choose among “constant”, “uniform”, “gaussian”, “lognormal”, “lin_corr”, “log_corr”).
parameters (dict, optional (default: {})) – Dictionary containing the properties of the weight distribution. Properties are as follow for the distributions
‘constant’: ‘value’
‘uniform’: ‘lower’, ‘upper’
‘gaussian’: ‘avg’, ‘std’
‘lognormal’: ‘position’, ‘scale’
noise_scale (class:int, optional (default: None)) – Scale of the multiplicative Gaussian noise that should be applied on the weights.
Note
If distribution and parameters are provided and the weights are set for the whole graph (elist is None), then the distribution properties will be kept as the new default for subsequent edges. That is, if new edges are created without specifying their weights, then these new weights will automatically be drawn from this previous distribution.
- property structure#
Object structuring the graph into specific groups.
Note
Points to
populationif the graph is aNetwork.
- to_file(filename, fmt='auto', separator=' ', secondary=';', attributes=None, notifier='@')[source]#
Save graph to file; options detailed below.
See also
nngt.lib.save_to_file()
- property type#
Type of the graph.
- class nngt.SpatialGraph(*args, **kwargs)[source]#
The detailed class that inherits from
Graphand implements additional properties to describe spatial graphs (i.e. graph where the structure is embedded in space.Initialize SpatialClass instance.
- Parameters
nodes (int, optional (default: 0)) – Number of nodes in the graph.
name (string, optional (default: “Graph”)) – The name of this
Graphinstance.weighted (bool, optional (default: True)) – Whether the graph edges have weight properties.
directed (bool, optional (default: True)) – Whether the graph is directed or undirected.
shape (
Shape, optional (default: None)) – Shape of the neurons’ environment (None leads to a square of side 1 cm)positions (
numpy.array(N, 2), optional (default: None)) – Positions of the neurons; if not specified and nodes is not 0, then neurons will be reparted at random inside theShapeobject of the instance.**kwargs (keyword arguments for
Graphor) –Shapeif no shape was given.
- Returns
self (
SpatialGraph)
- class nngt.Network(*args, **kwargs)[source]#
The detailed class that inherits from
Graphand implements additional properties to describe various biological functions and interact with the NEST simulator.Initializes
Networkinstance.- Parameters
nodes (int, optional (default: 0)) – Number of nodes in the graph.
name (string, optional (default: “Graph”)) – The name of this
Graphinstance.weighted (bool, optional (default: True)) – Whether the graph edges have weight properties.
directed (bool, optional (default: True)) – Whether the graph is directed or undirected.
copy_graph (
GraphObject, optional (default: None)) – An optionalGraphObjectto serve as base.population (
nngt.NeuralPop, (default: None)) – An object containing the neural groups and their properties: model(s) to use in NEST to simulate the neurons as well as their parameters.inh_weight_factor (float, optional (default: 1.)) – Factor to apply to inhibitory synapses, to compensate for example the strength difference due to timescales between excitatory and inhibitory synapses.
- Returns
self (
Network)
- class nngt.SpatialNetwork(*args, **kwargs)[source]#
Class that inherits from
NetworkandSpatialGraphto provide a detailed description of a real neural network in space, i.e. with positions and biological properties to interact with NEST.Initialize SpatialNetwork instance.
- Parameters
name (string, optional (default: “Graph”)) – The name of this
Graphinstance.weighted (bool, optional (default: True)) – Whether the graph edges have weight properties.
directed (bool, optional (default: True)) – Whether the graph is directed or undirected.
shape (
Shape, optional (default: None)) – Shape of the neurons’ environment (None leads to a square of side 1 cm)positions (
numpy.array, optional (default: None)) – Positions of the neurons; if not specified and nodes != 0, then neurons will be reparted at random inside theShapeobject of the instance.population (class:~nngt.NeuralPop, optional (default: None)) – Population from which the network will be built.
- Returns
self (
SpatialNetwork)