| Parameters: |
- network (
Graph or subclass) – The graph/network to plot.
- nsize (float, array of float or string, optional (default: “total-degree”)) – Size of the nodes as a percentage of the canvas length. Otherwise, it
can be a string that correlates the size to a node attribute among
“in/out/total-degree”, “in/out/total-strength”, or “betweenness”.
- ncolor (float, array of floats or string, optional (default: 0.5)) – Color of the nodes; if a float in [0, 1], position of the color in the
current palette, otherwise a string that correlates the color to a node
attribute among “in/out/total-degree”, “betweenness” or “group”.
- nshape (char, array of chars, or groups, optional (default: “o”)) – Shape of the nodes (see Matplotlib markers).
When using groups, they must be pairwise disjoint; markers will be
selected iteratively from the matplotlib default markers.
- nborder_color (char, float or array, optional (default: “k”)) – Color of the node’s border using predefined Matplotlib colors).
or floats in [0, 1] defining the position in the palette.
- nborder_width (float or array of floats, optional (default: 0.5)) – Width of the border in percent of canvas size.
- esize (float, str, or array of floats, optional (default: 0.5)) – Width of the edges in percent of canvas length. Available string values
are “betweenness” and “weight”.
- ecolor (str, char, float or array, optional (default: “k”)) – Edge color. If ecolor=”groups”, edges color will depend on the source
and target groups, i.e. only edges from and toward same groups will
have the same color.
- max_esize (float, optional (default: 5.)) – If a custom property is entered as esize, this normalizes the edge
width between 0. and max_esize.
- threshold (float, optional (default: 0.5)) – Size under which edges are not plotted.
- decimate_connections (int, optional (default: keep all connections)) – Plot only one connection every decimate_connections.
Use -1 to hide all edges.
- spatial (bool, optional (default: True)) – If True, use the neurons’ positions to draw them.
- restrict_sources (str, group, or list, optional (default: all)) – Only draw edges starting from a restricted set of source nodes.
- restrict_targets (str, group, or list, optional (default: all)) – Only draw edges ending on a restricted set of target nodes.
- restrict_nodes (str, group, or list, optional (default: plot all nodes)) – Only draw a subset of nodes.
- restrict_edges (list of edges, optional (default: all)) – Only draw a subset of edges.
- show_environment (bool, optional (default: True)) – Plot the environment if the graph is spatial.
- fast (bool, optional (default: False)) – Use a faster algorithm to plot the edges. Zooming on the drawing made
using this method leaves the size of the nodes and edges unchanged, it
is therefore not recommended when size consistency matters, e.g. for
some spatial representations.
- size (tuple of ints, optional (default: (600,600))) – (width, height) tuple for the canvas size (in px).
- dpi (int, optional (default: 75)) – Resolution (dot per inch).
- axis (matplotlib axis, optional (default: create new axis)) – Axis on which the network will be plotted.
- colorbar (bool, optional (default: False)) – Whether to display a colorbar for the node colors or not.
- cb_label (str, optional (default: None)) – A label for the colorbar.
- layout (str, optional (default: random or spatial positions)) – Name of a standard layout to structure the network. Available layouts
are: “circular” or “random”. If no layout is provided and the network
is spatial, then node positions will be used by default.
- show (bool, optional (default: True)) – Display the plot immediately.
- **kwargs (dict) – Optional keyword arguments including node_cmap to set the
nodes colormap (default is “magma” for continuous variables and
“Set1” for groups) and the boolean simple_nodes to make node
plotting faster.
|
|---|
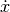 and
and 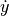 arrows should be
added to phase space. Requires additional ‘dotx’ and ‘doty’
arguments which are user defined functions to compute the
derivatives of x and x in time. These functions take 3
parameters, which are x, y, and time_dependent, where the
last parameter is a list of doubles associated to recordables
from the neuron model (see example for details). These recordables
must be declared in a time_dependent parameter.
arrows should be
added to phase space. Requires additional ‘dotx’ and ‘doty’
arguments which are user defined functions to compute the
derivatives of x and x in time. These functions take 3
parameters, which are x, y, and time_dependent, where the
last parameter is a list of doubles associated to recordables
from the neuron model (see example for details). These recordables
must be declared in a time_dependent parameter.