Plot module#
Functions for plotting graphs and graph properties.
The following features are provided:
basic graph plotting
plotting the distribution of some attribute over the graph
animation of some recorded activity
Content#
|
Class to plot the raster plot, firing-rate, and average trajectory in a 2D phase-space for a network activity. |
|
Class to plot the raster plot, firing-rate, and space-embedded spiking activity (neurons on the graph representation flash when spiking) in time. |
|
Plotting the betweenness distribution of a graph. |
|
Plot a chord diagram. |
Compare node attributes between two sets of nodes. |
|
|
For each node plot the value of reference_attributes against each of the other_attributes to check for correlations. |
|
Plotting the degree distribution of a graph. |
|
Draw a given graph/network. |
Return node attributes for a set of nodes. |
|
|
Draw a hive plot of the graph. |
|
Draw a given |
Return node attributes for a set of nodes. |
|
|
|
|
Details#
- class nngt.plot.Animation2d(source, multimeter, start=0.0, timewindow=None, trace=5.0, x='time', y='V_m', sort_neurons=None, network=None, interval=50, vector_field=False, **kwargs)[source]#
Class to plot the raster plot, firing-rate, and average trajectory in a 2D phase-space for a network activity.
Generate a SubplotAnimation instance to plot a network activity.
- Parameters:
source (tuple) – NEST gid of the ``spike_detector``(s) which recorded the network.
multimeter (tuple) – NEST gid of the ``multimeter``(s) which recorded the network.
timewindow (double, optional (default: None)) – Time window which will be shown for the spikes and self.second.
trace (double, optional (default: 5.)) – Interval of time (ms) over which the data is overlayed in red.
x (str, optional (default: “time”)) – Name of the x-axis variable (must be either “time” or the name of a NEST recordable in the multimeter).
y (str, optional (default: “V_m”)) – Name of the y-axis variable (must be either “time” or the name of a NEST recordable in the multimeter).
vector_field (bool, optional (default: False)) – Whether the
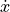 and
and 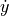 arrows should be
added to phase space. Requires additional ‘dotx’ and ‘doty’
arguments which are user defined functions to compute the
derivatives of x and x in time. These functions take 3
parameters, which are x, y, and time_dependent, where the
last parameter is a list of doubles associated to recordables
from the neuron model (see example for details). These recordables
must be declared in a time_dependent parameter.
arrows should be
added to phase space. Requires additional ‘dotx’ and ‘doty’
arguments which are user defined functions to compute the
derivatives of x and x in time. These functions take 3
parameters, which are x, y, and time_dependent, where the
last parameter is a list of doubles associated to recordables
from the neuron model (see example for details). These recordables
must be declared in a time_dependent parameter.sort_neurons (str or list, optional (default: None)) – Sort neurons using a topological property (“in-degree”, “out-degree”, “total-degree” or “betweenness”), an activity-related property (“firing_rate”, ‘B2’) or a user-defined list of sorted neuron ids. Sorting is performed by increasing value of the sort_neurons property from bottom to top inside each group.
**kwargs (dict, optional (default: {})) – Optional arguments such as ‘make_rate’, ‘num_xarrows’, ‘num_yarrows’, ‘dotx’, ‘doty’, ‘time_dependent’, ‘recordables’, ‘arrow_scale’.
- class nngt.plot.AnimationNetwork(source, network, resolution=1.0, start=0.0, timewindow=None, trace=5.0, show_spikes=False, sort_neurons=None, decimate_connections=False, interval=50, repeat=True, resting_size=None, active_size=None, **kwargs)[source]#
Class to plot the raster plot, firing-rate, and space-embedded spiking activity (neurons on the graph representation flash when spiking) in time.
Generate a SubplotAnimation instance to plot a network activity.
- Parameters:
source (tuple) – NEST gid of the ``spike_detector``(s) which recorded the network.
network (
SpatialNetwork) – Network embedded in space to plot the actvity of the neurons in space.resolution (double, optional (default: None)) – Time resolution of the animation.
timewindow (double, optional (default: None)) – Time window which will be shown for the spikes and self.second.
trace (double, optional (default: 5.)) – Interval of time (ms) over which the data is overlayed in red.
show_spikes (bool, optional (default: True)) – Whether a spike trajectory should be displayed on the network.
sort_neurons (str or list, optional (default: None)) – Sort neurons using a topological property (“in-degree”, “out-degree”, “total-degree” or “betweenness”), an activity-related property (“firing_rate”, ‘B2’) or a user-defined list of sorted neuron ids. Sorting is performed by increasing value of the sort_neurons property from bottom to top inside each group.
**kwargs (dict, optional (default: {})) – Optional arguments such as ‘make_rate’, or all arguments for the
nngt.plot.draw_network().
- nngt.plot.betweenness_distribution(network, btype='both', weights=False, nodes=None, logx=False, logy=False, num_nbins=None, num_ebins=None, axes=None, colors=None, norm=False, legend_location='right', title=None, show=True, **kwargs)[source]#
Plotting the betweenness distribution of a graph.
Changed in version 2.5.0: Added title argument.
- Parameters:
graph (
Graphor subclass) – the graph to analyze.btype (string, optional (default: “both”)) – type of betweenness to display (“node”, “edge” or “both”)
weights (bool or str, optional (default: binary edges)) – Whether edge weights should be considered; if
NoneorFalsethen use binary edges; ifTrue, uses the ‘weight’ edge attribute, otherwise uses any valid edge attribute required.nodes (list or numpy.array of ints, optional (default: all nodes)) – Restrict the distribution to a set of nodes (taken into account only for the node attribute).
logx (bool, optional (default: False)) – use log-spaced bins.
logy (bool, optional (default: False)) – use logscale for the degree count.
num_nbins (int or ‘auto’, optional (default: None):) – Number of bins used to sample the node distribution. Defaults to max(num_nodes / 50., 10).
num_ebins (int or ‘auto’, optional (default: None):) – Number of bins used to sample the edge distribution. Defaults to max(num_edges / 500., 10) (‘auto’ method will be slow).
axes (list of
matplotlib.axis.Axis, optional (default: new ones)) – Axes which should be used to plot the histogram, if None, new ones are created.legend_location (str, optional (default; ‘right’)) – Location of the legend.
title (str, optional (default: auto-generated)) – Title of the axis.
show (bool, optional (default: True)) – Show the Figure right away if True, else keep it warm for later use.
- nngt.plot.chord_diagram(network, weights=True, names=None, order=None, width=0.1, pad=2.0, gap=0.03, chordwidth=0.7, axis=None, colors=None, cmap=None, alpha=0.7, use_gradient=False, chord_colors=None, start_at=0, extent=360, directed=None, show=False, **kwargs)[source]#
Plot a chord diagram.
- Parameters:
network (a
nngt.Graphobject) – Network used to plot the chord diagram.weights (bool or str, optional (default: ‘weight’ attribute)) – Weights used to plot the connections.
names (str or list of str, optional (default: no names)) – Names of the nodes that will be displayed, either a node attribute or a custom list (must be ordered following the nodes’ indices).
order (list, optional (default: order of the matrix entries)) – Order in which the arcs should be placed around the trigonometric circle.
width (float, optional (default: 0.1)) – Width/thickness of the ideogram arc.
pad (float, optional (default: 2)) – Distance between two neighboring ideogram arcs. Unit: degree.
gap (float, optional (default: 0.03)) – Distance between the arc and the beginning of the cord.
chordwidth (float, optional (default: 0.7)) – Position of the control points for the chords, controlling their shape.
axis (matplotlib axis, optional (default: new axis)) – Matplotlib axis where the plot should be drawn.
colors (list, optional (default: from cmap)) – List of user defined colors or floats.
cmap (str or colormap object (default: viridis)) – Colormap that will be used to color the arcs and chords by default. See chord_colors to use different colors for chords.
alpha (float in [0, 1], optional (default: 0.7)) – Opacity of the chord diagram.
use_gradient (bool, optional (default: False)) – Whether a gradient should be use so that chord extremities have the same color as the arc they belong to.
chord_colors (str, or list of colors, optional (default: None)) – Specify color(s) to fill the chords differently from the arcs. When the keyword is not used, chord colors default to the colomap given by colors. Possible values for chord_colors are:
a single color (do not use an RGB tuple, use hex format instead), e.g. “red” or “#ff0000”; all chords will have this color
a list of colors, e.g.
["red", "green", "blue"], one per node (in this case, RGB tuples are accepted as entries to the list). Each chord will get its color from its associated source node, or from both nodes if use_gradient is True.
start_at (float, optional (default : 0)) – Location, in degrees, where the diagram should start on the unit circle. Default is to start at 0 degrees, i.e. (x, y) = (1, 0) or 3 o’clock), and move counter-clockwise
extent (float, optional (default : 360)) – The angular aperture, in degrees, of the diagram. Default is to use the whole circle, i.e. 360 degrees, but in some cases it can be useful to use only a part of it.
directed (bool, optional (default: same as network)) – Whether the chords should be directed with one part of each arc dedicated to outgoing chords and the other to incoming ones.
show (bool, optional (default: False)) – Whether the plot should be displayed immediately via an automatic call to plt.show().
kwargs (keyword arguments) – Available kwargs are:
Name
Type
Purpose and possible values
fontcolor
str or list
Color of the names
fontsize
int
Size of the font for names
rotate_names
(list of) bool(s)
Rotate names by 90°
sort
str
Either “size” or “distance”
zero_entry_size
float
Size of zero-weight reciprocal
- nngt.plot.compare_population_attributes(network, attributes, nodes=None, reference_nodes=None, num_bins='auto', reference_color='gray', title=None, logx=False, logy=False, show=True, **kwargs)[source]#
Compare node attributes between two sets of nodes. Since number of nodes can vary, normalized distributions are used.
- Parameters:
network (
Graph) – The graph where the nodes belong.attributes (str or list) – Attributes which should be returned, among: * “betweenness” * “clustering” * “in-degree”, “out-degree”, “total-degree” * “subgraph_centrality” * “b2” (requires NEST) * “firing_rate” (requires NEST)
nodes (list, optional (default: all nodes)) – Nodes for which the attributes should be returned.
reference_nodes (list, optional (default: all nodes)) – Reference nodes for which the attributes should be returned in order to compare with nodes.
num_bins (int or list, optional (default: ‘auto’)) – Number of bins to plot the distributions. If only one int is provided, it is used for all attributes, otherwize a list containing one int per attribute in attributes is required. Defaults to unsupervised Bayesian blocks method.
logx (bool or list, optional (default: False)) – Use log-spaced bins.
logy (bool or list, optional (default: False)) – use logscale for the node count.
- nngt.plot.correlation_to_attribute(network, reference_attribute, other_attributes, attribute_type='node', nodes=None, edges=None, fig=None, title=None, show=True)[source]#
For each node plot the value of reference_attributes against each of the other_attributes to check for correlations.
Changed in version 2.0: Added fig argument.
- Parameters:
network (
Graph) – The graph where the nodes belong.reference_attribute (str or array-like) – Attribute which should serve as reference, among:
“betweenness”
“clustering”
“in-degree”, “out-degree”, “total-degree”
“in-strength”, “out-strength”, “total-strength”
“subgraph_centrality”
“b2” (requires NEST)
“firing_rate” (requires NEST)
a custom array of values, in which case one entry per node in nodes is required.
other_attributes (str or list) – Attributes that will be compared to the reference.
attribute_type (str, optional (default: ‘node’)) – Whether we are dealing with ‘node’ or ‘edge’ attributes
nodes (list, optional (default: all nodes)) – Nodes for which the attributes should be returned.
edges (list, optional (default: all edges)) – Edges for which the attributes should be returned.
fig (
matplotlib.figure.Figure, optional (default: new Figure)) – Figure to which the plot should be added.title (str, optional (default: automatic).) – Custom title, use “” to remove the automatic title.
show (bool, optional (default: True)) – Whether the plot should be displayed immediately.
- nngt.plot.degree_distribution(network, deg_type='total', nodes=None, num_bins='doane', weights=False, logx=False, logy=False, axis=None, colors=None, norm=False, show=True, title=None, **kwargs)[source]#
Plotting the degree distribution of a graph.
Changed in version 2.5.0: Removed unused axis_num argument.
- Parameters:
graph (
Graphor subclass) – The graph to analyze.deg_type (string or N-tuple, optional (default: “total”)) – Type of degree to consider (“in”, “out”, or “total”)
nodes (list or numpy.array of ints, optional (default: all nodes)) – Restrict the distribution to a set of nodes.
num_bins (str, int or N-tuple, optional (default: ‘doane’):) – Number of bins used to sample the distribution. Defaults to ‘doane’. Use to ‘auto’ for numpy automatic selection or ‘bayes’ for unsupervised Bayesian blocks method.
weights (bool or str, optional (default: binary edges)) – Whether edge weights should be considered; if
NoneorFalsethen use binary edges; ifTrue, uses the ‘weight’ edge attribute, otherwise uses any valid edge attribute required.logx (bool, optional (default: False)) – Use log-spaced bins.
logy (bool, optional (default: False)) – Use logscale for the degree count.
axis (
matplotlib.axes.Axesinstance, optional (default: new one)) – Axis which should be used to plot the histogram, if None, a new one is created.colors ((list of) matplotlib colors, optional (default: from palette)) – Colors associated to each degree type.
title (str, optional (default: no title)) – Title of the axis.
show (bool, optional (default: True)) – Show the Figure right away if True, else keep it warm for later use.
**kwargs (keyword arguments for
matplotlib.axes.Axes.bar().)
- nngt.plot.draw_network(network, nsize='total-degree', ncolor=None, nshape='o', esize=None, ecolor='k', curved_edges=False, threshold=0.5, decimate_connections=None, spatial=True, restrict_sources=None, restrict_targets=None, restrict_nodes=None, restrict_edges=None, show_environment=True, fast=False, size=(600, 600), xlims=None, ylims=None, dpi=75, axis=None, colorbar=False, cb_label=None, layout=None, show=False, **kwargs)[source]#
Draw a given graph/network.
- Parameters:
network (
Graphor subclass) – The graph/network to plot.nsize (float, array of float or string, optional (default: “total-degree”)) – Size of the nodes as a percentage of the canvas length. Otherwise, it can be a string that correlates the size to a node attribute among “in/out/total-degree”, “in/out/total-strength”, or “betweenness”.
ncolor (float, array of floats or string, optional) – Color of the nodes; if a float in [0, 1], position of the color in the current palette, otherwise a string that correlates the color to a node attribute or “in/out/total-degree”, “betweenness” and “group”. Default to red or one color per group in the graph if not specified.
nshape (char, array of chars, or groups, optional (default: “o”)) – Shape of the nodes (see Matplotlib markers). When using groups, they must be pairwise disjoint; markers will be selected iteratively from the matplotlib default markers.
nborder_color (char, float or array, optional (default: “k”)) – Color of the node’s border using predefined Matplotlib colors). or floats in [0, 1] defining the position in the palette.
nborder_width (float or array of floats, optional (default: 0.5)) – Width of the border in percent of canvas size.
esize (float, str, or array of floats, optional (default: 0.5)) – Width of the edges in percent of canvas length. Available string values are “betweenness” and “weight”.
ecolor (str, char, float or array, optional (default: “k”)) – Edge color. If ecolor=”groups”, edges color will depend on the source and target groups, i.e. only edges from and toward same groups will have the same color.
curved_edges (bool, optional (default: False)) – Whether the edges should be curved or straight.
threshold (float, optional (default: 0.5)) – Size under which edges are not plotted.
decimate_connections (int, optional (default: keep all connections)) – Plot only one connection every decimate_connections. Use -1 to hide all edges.
spatial (bool, optional (default: True)) – If True, use the neurons’ positions to draw them.
restrict_sources (str, group, or list, optional (default: all)) – Only draw edges starting from a restricted set of source nodes.
restrict_targets (str, group, or list, optional (default: all)) – Only draw edges ending on a restricted set of target nodes.
restrict_nodes (str, group, or list, optional (default: plot all nodes)) – Only draw a subset of nodes.
restrict_edges (list of edges, optional (default: all)) – Only draw a subset of edges.
show_environment (bool, optional (default: True)) – Plot the environment if the graph is spatial.
fast (bool, optional (default: False)) – Use a faster algorithm to plot the edges. Zooming on the drawing made using this method leaves the size of the nodes and edges unchanged, it is therefore not recommended when size consistency matters, e.g. for some spatial representations.
size (tuple of ints, optional (default: (600,600))) – (width, height) tuple for the canvas size (in px).
dpi (int, optional (default: 75)) – Resolution (dot per inch).
axis (matplotlib axis, optional (default: create new axis)) – Axis on which the network will be plotted.
colorbar (bool, optional (default: False)) – Whether to display a colorbar for the node colors or not.
cb_label (str, optional (default: None)) – A label for the colorbar.
layout (str, optional (default: random or spatial positions)) – Name of a standard layout to structure the network. Available layouts are: “circular” or “random”. If no layout is provided and the network is spatial, then node positions will be used by default.
show (bool, optional (default: True)) – Display the plot immediately.
**kwargs (dict) – Optional keyword arguments.
Name
Type
Purpose and possible values
node_cmap
str
“magma” for continuous variables and “Set1” for groups)
title
str
Title of the plot
max_*
float
Maximum value for nsize or esize
min_*
float
Minimum value for nsize or esize
nalpha
float
Node opacity in [0, 1]`, default 1
ealpha
float
Edge opacity, default 0.5
*border_color
color
or edges (e). Default to black.
*border_width
float
(e). Default to .5 for nodes and .3 for edges (if fast is False).
simple_nodes
bool
are always the same size) or patches (change size with zoom).
- nngt.plot.edge_attributes_distribution(network, attributes, edges=None, num_bins='auto', logx=False, logy=False, norm=False, title=None, axtitles=None, colors=None, axes=None, show=True, **kwargs)[source]#
Return node attributes for a set of nodes.
Changed in version 2.5.0: Added axtitles and axes arguments.
- Parameters:
network (
Graph) – The graph where the nodes belong.attributes (str or list) – Attributes which should be returned (e.g. “betweenness”, “delay”, “weight”).
edges (list, optional (default: all edges)) – Edges for which the attributes should be returned.
num_bins (int or list, optional (default: ‘auto’)) – Number of bins to plot the distributions. If only one int is provided, it is used for all attributes, otherwise a list containing one int per attribute in attributes is required. Defaults to unsupervised Bayesian blocks method.
logx (bool or list, optional (default: False)) – Use log-spaced bins.
logy (bool or list, optional (default: False)) – use logscale for the node count.
norm (bool, optional (default: False)) – Whether the histogram should be normed such that the sum of the counts is 1.
title (str, optional (default: no title)) – Title of the figure.
axtitles (list of str, optional (default: auto-generated)) – Titles of the axes. Use “” or False to turn them of.
colors ((list of) matplotlib colors, optional (default: from palette)) – Colors associated to each degree type.
axes (list of
matplotlib.axis.Axis, optional (default: new ones)) – Axess which should be used to plot the histograms, if None, a new axis is created for each attribute.show (bool, optional (default: True)) – Show the Figure right away if True, else keep it warm for later use.
**kwargs (keyword arguments for
matplotlib.axes.Axes.bar().)
- nngt.plot.hive_plot(network, radial, axes=None, axes_bins=None, axes_range=None, axes_angles=None, axes_labels=None, axes_units=None, intra_connections=True, highlight_nodes=None, highlight_edges=None, nsize=None, esize=None, max_nsize=10, max_esize=1, axes_colors=None, edge_colors=None, edge_alpha=0.05, nborder_color='k', nborder_width=0.2, show_names=True, show_circles=False, axis=None, tight=True, show=False)[source]#
Draw a hive plot of the graph.
Note
For directed networks, the direction of intra-axis connections is counter-clockwise. For inter-axes connections, the default edge color is closest to the color of the source group (i.e. from a red group to a blue group, edge color will be a reddish violet , while from blue to red, it will be a blueish violet).
- Parameters:
network (
Graph) – Graph to plot.radial (str, list of str or array-like) – Values that will be used to place the nodes on the axes. Either one identical property is used for all axes (traditional hive plot) or one radial coordinate per axis is used (custom hive plot). If radial is a string or a list of strings, then these must correspond to the names of node attributes stored in the graph.
axes (str, or list of str, optional (default: one per radial coordinate)) – Name of the attribute(s) that will be used to make each of the axes (i.e. each group of nodes). This can be either “groups” if the graph has a structure or is a
Network, a list of (Meta)Group names, or any (list of) node attribute(s). If a single node attribute is used, axes_bins must be provided to make one axis for each range of values. If there are multiple radial coordinates, then leaving axes blanck will plot all nodes on each of the axes (one per radial coordinate).axes_bins (int or array-like, optional (default: all nodes on each axis)) – Required if there is a single radial coordinate and a single axis entry: provides the bins that will be used to separate the nodes into groups (one per axis). For N axes, there must therefore be N + 1 entries in axes_bins, or axis_bins must be equal to N, in which case the nodes are separated into N evenly sized bins.
axes_units (str, optional) – Units used to scale the axes. Either “native” to have them scaled between the minimal and maximal radial coordinates among all axes, “rank”, to use the min and max ranks of the nodes on all axes, or “normed”, to have each axis go from zero (minimal local radial coordinate) to one (maximal local radial coordinate). “native” is the default if there is a single radial coordinate, “normed” is the default for multiple coordinates.
axes_angles (list of angles, optional (default: automatic)) – Angles for each of the axes, by increasing degree. If intra_connections is True, then angles of duplicate axes must be adjacent, e.g.
[a1, a1bis, a2, a2bis, a3, a3bis].axes_labels (str or list of str, optional) – Label of each axis. For binned axes, it can be automatically formatted via the three entries
{name},{start},{stop}. E.g. “{name} in [{start}, {stop}]” would give “CC in [0, 0.2]” for a first axis and “CC in [0.2, 0.4]” for a second axis.intra_connections (bool, optional (default: True)) – Show connections between nodes belonging to the same axis. If true, then each axis is duplicated to display intra-axis connections.
highlight_nodes (list of nodes, optional (default: all nodes)) – Highlight a subset of nodes and their connections, all other nodes and connections will be gray.
highlight_edges (list of edges, optional (default: all edges)) – Highlight a subset of edges; all other connections will be gray.
nsize (float, str, or array-like, optional (default: automatic)) – Size of the nodes on the axes. Either a fixed size, the name of a node attribute, or a list of user-defined values.
esize (float or str, optional (default: 1)) – Size of the edges. Either a fixed size or the name of an edge attribute.
max_nsize (float, optional (default: 10)) – Maximum node size if nsize is an attribute or a list of user-defined values.
max_esize (float, optional (default: 1)) – Maximum edge size if esize is an attribute.
axes_colors (valid matplotlib color/colormap, optional (default: Set1)) – Color associated to each axis.
nborder_color (matplotlib color, optional (default: “k”)) – Color of the node’s border. or floats in [0, 1] defining the position in the palette.
nborder_width (float, optional (default: 0.2)) – Width of the border.
edge_colors (valid matplotlib color/colormap, optional (default: auto)) – Color of the edges. By default it is the intermediate color between two axes colors. To provide custom colors, they must be provided as a dictionnary of axes edges
{(0, 0): "r", (0, 1): "g", (1, 0): "b"}with default color being black.edge_alpha (float, optional (default: 0.05)) – Edge opacity.
show_names (bool, optional (default: True)) – Show axes names and properties.
show_circles (bool, optional (default: False)) – Show the circles associated to the maximum value of each axis.
axis (matplotlib axis, optional (default: create new axis)) – Axis on which the network will be plotted.
tight (bool, optional (default: True)) – Set figure layout to tight (set to False if plotting multiple axes on a single figure).
show (bool, optional (default: True)) – Display the plot immediately.
- nngt.plot.library_draw(network, nsize='total-degree', ncolor=None, nshape='o', nborder_color='k', nborder_width=0.5, esize=1.0, ecolor='k', ealpha=0.5, curved_edges=False, threshold=0.5, decimate_connections=None, spatial=True, restrict_sources=None, restrict_targets=None, restrict_nodes=None, restrict_edges=None, show_environment=True, size=(600, 600), xlims=None, ylims=None, dpi=75, axis=None, colorbar=False, show_labels=False, layout=None, show=False, **kwargs)[source]#
Draw a given
Graphusing the underlying library’s drawing functions.New in version 2.0.
Warning
When using igraph or graph-tool, if you want to use the axis argument, then you must first switch the matplotlib backend to its cairo version using e.g.
plt.switch_backend("Qt5Cairo")if your normal backend is Qt5 (“Qt5Agg”).- Parameters:
network (
Graphor subclass) – The graph/network to plot.nsize (float, array of float or string, optional (default: “total-degree”)) – Size of the nodes as a percentage of the canvas length. Otherwise, it can be a string that correlates the size to a node attribute among “in/out/total-degree”, or “betweenness”.
ncolor (float, array of floats or string, optional (default: 0.5)) – Color of the nodes; if a float in [0, 1], position of the color in the current palette, otherwise a string that correlates the color to a node attribute or “in/out/total-degree”, “betweenness” and “group”. Default to red or one color per group in the graph if not specified.
nshape (char, array of chars, or groups, optional (default: “o”)) – Shape of the nodes (see Matplotlib markers). When using groups, they must be pairwise disjoint; markers will be selected iteratively from the matplotlib default markers.
nborder_color (char, float or array, optional (default: “k”)) – Color of the node’s border using predefined Matplotlib colors). or floats in [0, 1] defining the position in the palette.
nborder_width (float or array of floats, optional (default: 0.5)) – Width of the border in percent of canvas size.
esize (float, str, or array of floats, optional (default: 0.5)) – Width of the edges in percent of canvas length. Available string values are “betweenness” and “weight”.
ecolor (str, char, float or array, optional (default: “k”)) – Edge color. If ecolor=”groups”, edges color will depend on the source and target groups, i.e. only edges from and toward same groups will have the same color.
threshold (float, optional (default: 0.5)) – Size under which edges are not plotted.
decimate_connections (int, optional (default: keep all connections)) – Plot only one connection every decimate_connections. Use -1 to hide all edges.
spatial (bool, optional (default: True)) – If True, use the neurons’ positions to draw them.
restrict_sources (str, group, or list, optional (default: all)) – Only draw edges starting from a restricted set of source nodes.
restrict_targets (str, group, or list, optional (default: all)) – Only draw edges ending on a restricted set of target nodes.
restrict_nodes (str, group, or list, optional (default: plot all nodes)) – Only draw a subset of nodes.
restrict_edges (list of edges, optional (default: all)) – Only draw a subset of edges.
show_environment (bool, optional (default: True)) – Plot the environment if the graph is spatial.
size (tuple of ints, optional (default: (600, 600))) – (width, height) tuple for the canvas size (in px).
dpi (int, optional (default: 75)) – Resolution (dot per inch).
colorbar (bool, optional (default: False)) – Whether to display a colorbar for the node colors or not.
axis (matplotlib axis, optional (default: create new axis)) – Axis on which the network will be plotted.
layout (str, optional (default: library-dependent or spatial positions)) – Name of a standard layout to structure the network. Available layouts are: “circular”, “spring-block”, “random”. If no layout is provided and the network is spatial, then node positions will be used by default.
show (bool, optional (default: True)) – Display the plot immediately.
**kwargs (dict) – Optional keyword arguments.
Name
Type
Purpose and possible values
node_cmap
str
“magma” for continuous variables and “Set1” for groups)
title
str
Title of the plot
max_*
float
Maximum value for nsize or esize
min_*
float
Minimum value for nsize or esize
annotate
bool
Use annotations to show node information (default: True)
annotations
str or list
such as a node attribute or a list of values. (default: node id)
- nngt.plot.node_attributes_distribution(network, attributes, nodes=None, num_bins='auto', logx=False, logy=False, norm=False, title=None, axtitles=None, colors=None, axes=None, show=True, **kwargs)[source]#
Return node attributes for a set of nodes.
Changed in version 2.5.0: Added axtitles and axes arguments.
- Parameters:
network (
Graph) – The graph where the nodes belong.attributes (str or list) – Attributes which should be returned, among: * any user-defined node attribute * “betweenness” * “clustering” * “closeness” * “in-degree”, “out-degree”, “total-degree” * “subgraph_centrality” * “b2” (requires NEST) * “firing_rate” (requires NEST)
nodes (list, optional (default: all nodes)) – Nodes for which the attributes should be returned.
num_bins (int or list, optional (default: ‘auto’)) – Number of bins to plot the distributions. If only one int is provided, it is used for all attributes, otherwise a list containing one int per attribute in attributes is required. Defaults to unsupervised Bayesian blocks method.
logx (bool or list, optional (default: False)) – Use log-spaced bins.
logy (bool or list, optional (default: False)) – use logscale for the node count.
norm (bool, optional (default: False)) – Whether the histogram should be normed such that the sum of the counts is 1.
title (str, optional (default: no title)) – Title of the figure.
axtitles (list of str, optional (default: auto-generated)) – Titles of the axes. Use “” or False to turn them of.
colors ((list of) matplotlib colors, optional (default: from palette)) – Colors associated to each degree type.
axes (list of
matplotlib.axis.Axis, optional (default: new ones)) – Axess which should be used to plot the histograms, if None, a new axis is created for each attribute.show (bool, optional (default: True)) – Show the Figure right away if True, else keep it warm for later use.
**kwargs (keyword arguments for
matplotlib.axes.Axes.bar().)